Research
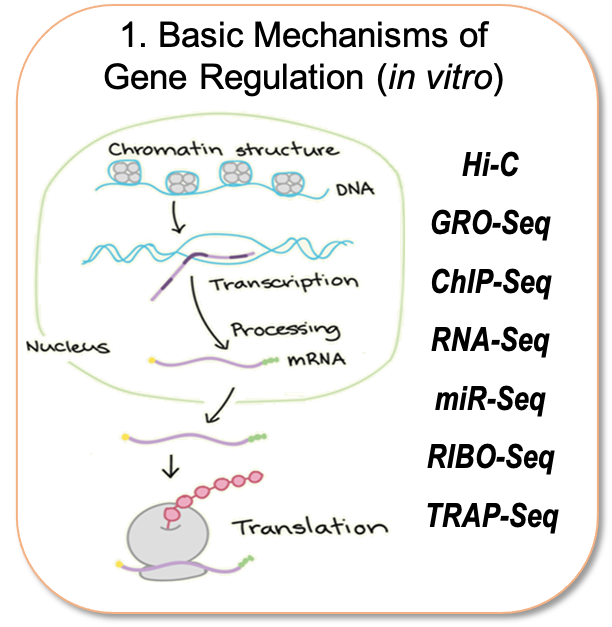
Our research group is interested in understanding how different cell types associated with coronary artery disease (CAD), namely those of the aortic wall, liver and adipose tissue contribute to disease pathogenesis. Towards this goal, we follow three lines of research outlined below (Figure). Especially, we are interested in understanding the regulatory code that drives cell type- and disease- specific gene expression which resides in distal elements called enhancers. To identify and characterize gene regulatory processes at the level of chromatin organization and transcription, our lab has gained extensive experience in a wide range of state-of-the-art sequencing methods that some are only mastered by a limited number of labs (e.g. Hi-C, GRO-Seq and snATAC-Seq). This has allowed us to make vital contributions to the field of vascular biology and provide genome-wide annotation of non-coding RNAs that are regulated by inflammation and atherosclerosis.
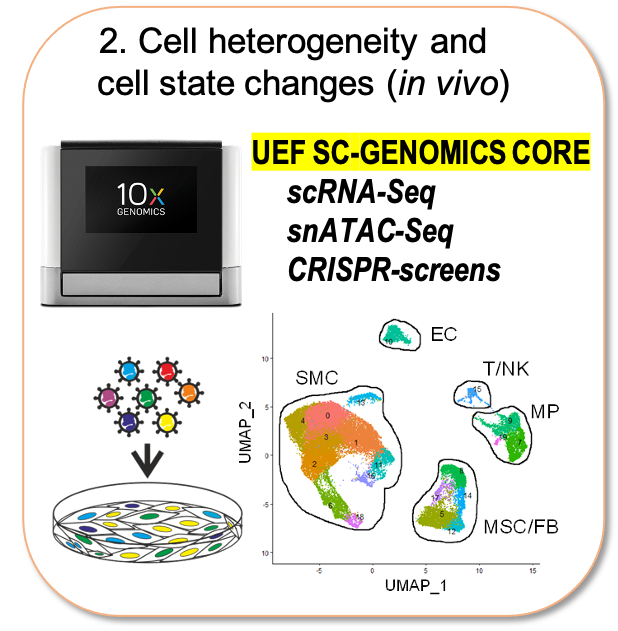
Recently the field has been revolutionized by the emergence of single cell technologies and our lab has been responsible for setting up this technology at the University of Eastern Finland (10XGenomics Chromium microfluidics system). The single cell genomics core at UEF now offers consultancy and training to other research groups and is organized under the umbrella of Biocenter Finland Single Cell Omics Platform. In our own research, we use this technology to characterize cell state changes associated with the progression of atherosclerosis across disease relevant tissues in mouse models and human patient samples. Unveiling the key regulatory factors and networks driving their pathological transformation could improve early detection and provide deeper understanding of the disease process.
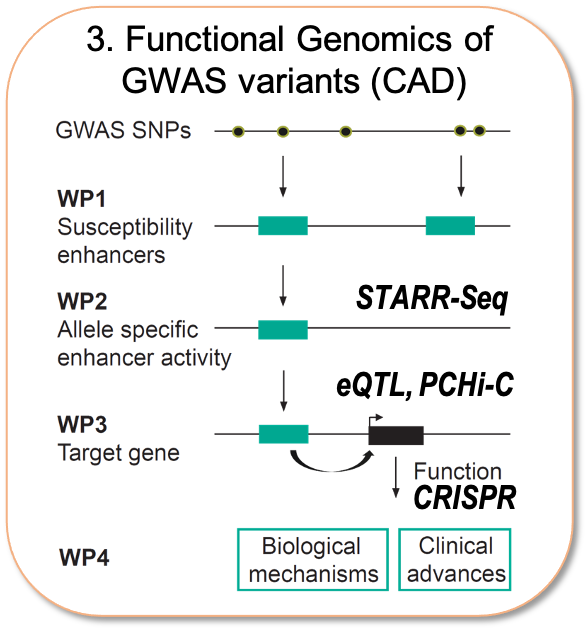
As the majority of the risk variants for cardiometabolic diseases are located within enhancers, we believe that understanding the genetic basis of disease requires the functional characterization of the non-coding variants in terms of allele-specific activity and target gene identification. To achieve this, my lab has recently set up massively parallel reporter assays (STARR-Seq) and CRISRP/Cas9-based screening methods and integrated as part of extensive international collaboration networks allowing access to unique patient cohorts for cell- and tissue- specific eQTL information. Ultimately, we wish to understand the relative contributions of distinct cell types in CAD and functionally characterize the genes influenced by non-coding variants.