Andrius Penkauskas: Addressing the infamous “So what” question
Scientific research is a formidable journey, often requiring months or even years to transform an initial idea into a tangible outcome. The process demands immense dedication, involving the navigation of complex problems, meticulous review and analysis of existing literature, scrutiny of data, and the development of code line by line. Each of these steps not only deepens one’s understanding but also illuminates the intricacies of the subject at hand. In time, the specific issues one seeks to address may become crystal clear, at least to the researcher himself.
Yet, a crucial question remains: does this clarity extend beyond your own understanding and resonate with the broader public?
Why the “So What?” Question Matters
To illustrate this challenge, imagine a scenario: you are discussing your research with someone entirely unfamiliar with your field. After offering a detailed and passionate explanation, you are met with a simple yet profound question: “So what?” This inquiry, though disarmingly straightforward, is entirely legitimate. Consider the title of one of my oral presentations at Hydrocephalus 2025 World Congress: “Exploring the Role of Lateral Ventricles’ Geometric Features in Predicting Extracellular Deposition of Amyloid-β Peptides in Patients with Idiopathic Normal Pressure Hydrocephalus.” To many, this may sound perplexing, perhaps even like a jumble of technical jargon.
Therefore, I invite you to join me as I attempt to answer the crucial “So what?” question and bridge the gap between specialized research and its broader significance in the context of my own work.
The Challenge of Distinguishing iNPH
My research focuses on Idiopathic Normal Pressure Hydrocephalus (iNPH), a subtype of dementia distinguished by its potential reversibility through shunt surgery. Unlike most other dementias, iNPH offers hope for significant clinical improvement. However, this optimism is tempered by a significant challenge: the presence of comorbid neurodegenerative conditions such as Alzheimer’s disease (AD) can undermine the effectiveness of surgical intervention.
Thus, accurately distinguishing iNPH from neurodegenerative forms, particularly NPH with Alzheimer’s pathology (NPH-AD), is crucial to avoid ineffective treatments and unnecessary patient hardship. This distinction, however, is far from straightforward because iNPH and NPH-AD often share overlapping pathological and clinical features.
The amyloid hypothesis of Alzheimer’s disease has long been the predominant theory, suggesting that Alzheimer’s disease is caused by the accumulation of amyloid beta protein (Aβ) in the brain, leading to neuronal toxicity in the central nervous system. The relationship between amyloid pathology observed in iNPH patients’ brain biopsy samples and the response to shunt placement has been documented, highlighting the significance of this pathology in guiding iNPH treatment decisions. However, the ground truth of amyloid pathology is acquired utilizing a biopsy sample, in other words, a small sample of the brain.
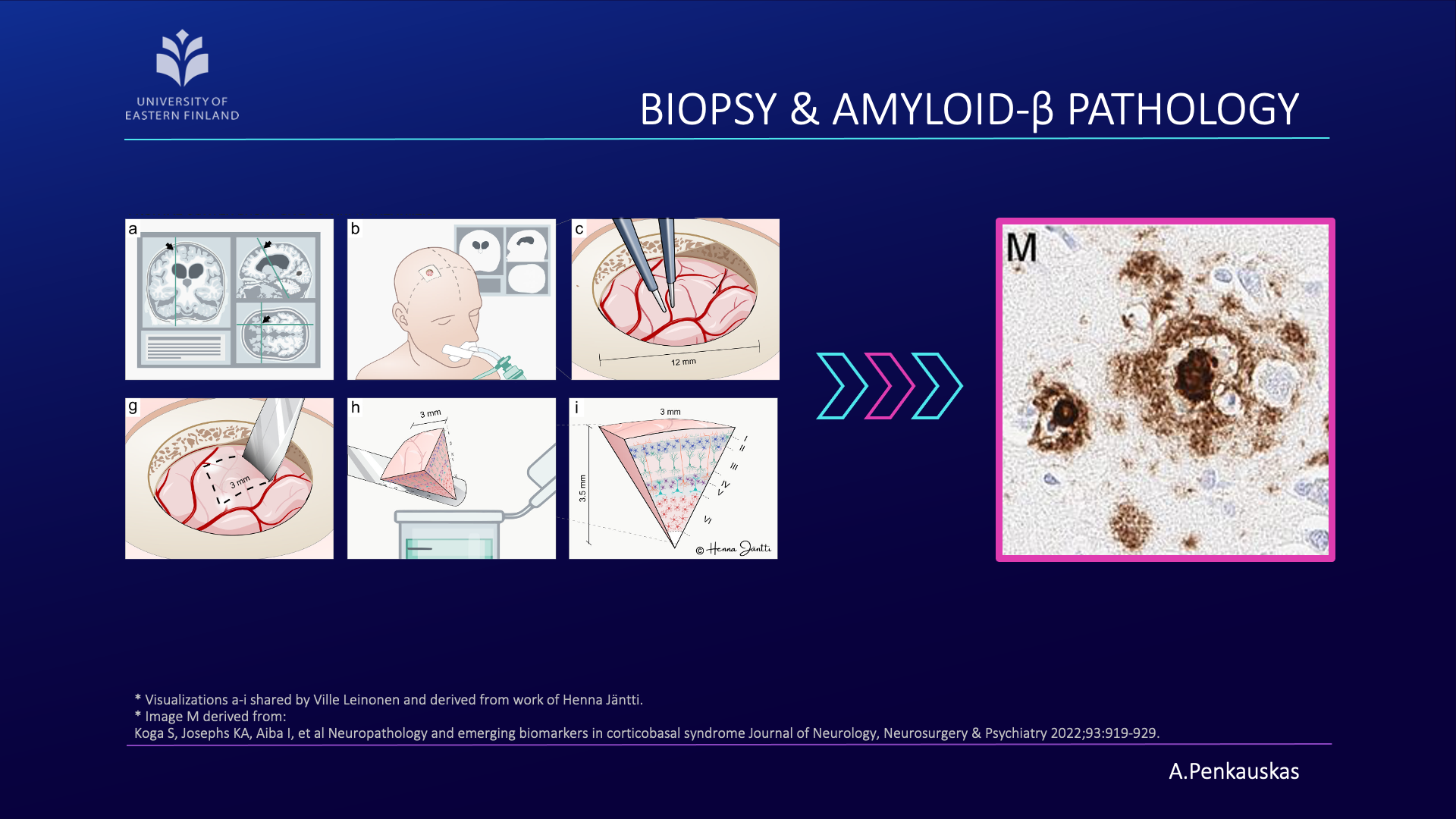
Towards Less Invasive Solutions
The invasive nature of a biopsy warrants its limited application. Due to this reason, my research effort was focused on developing a less invasive pipeline that would enable the prediction of amyloid pathology in iNPH patients.

Have we succeeded? In part, yes; the results are promising. However, to ensure our approach is robust and applicable across diverse patient populations, further validation with larger datasets is essential.
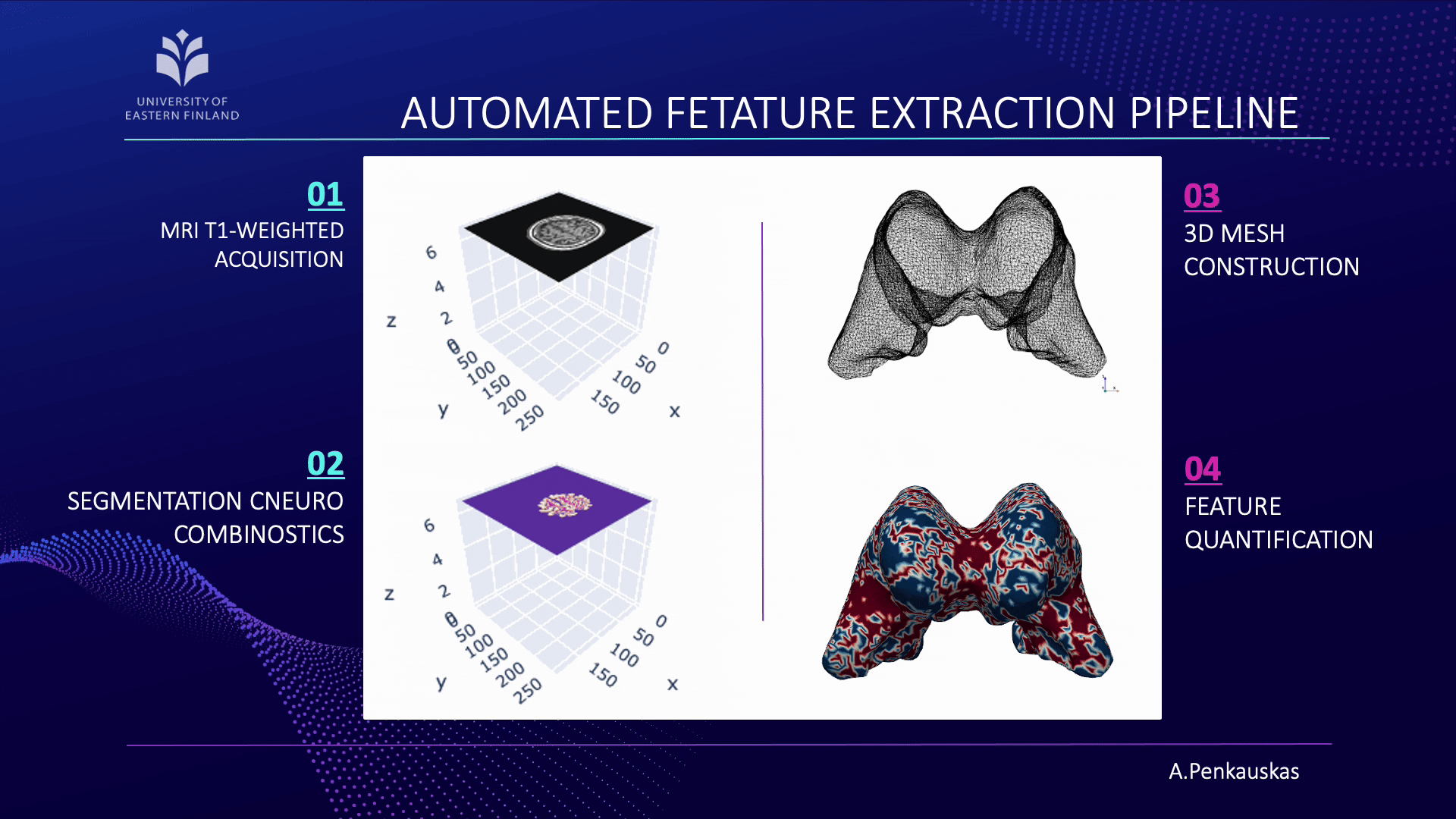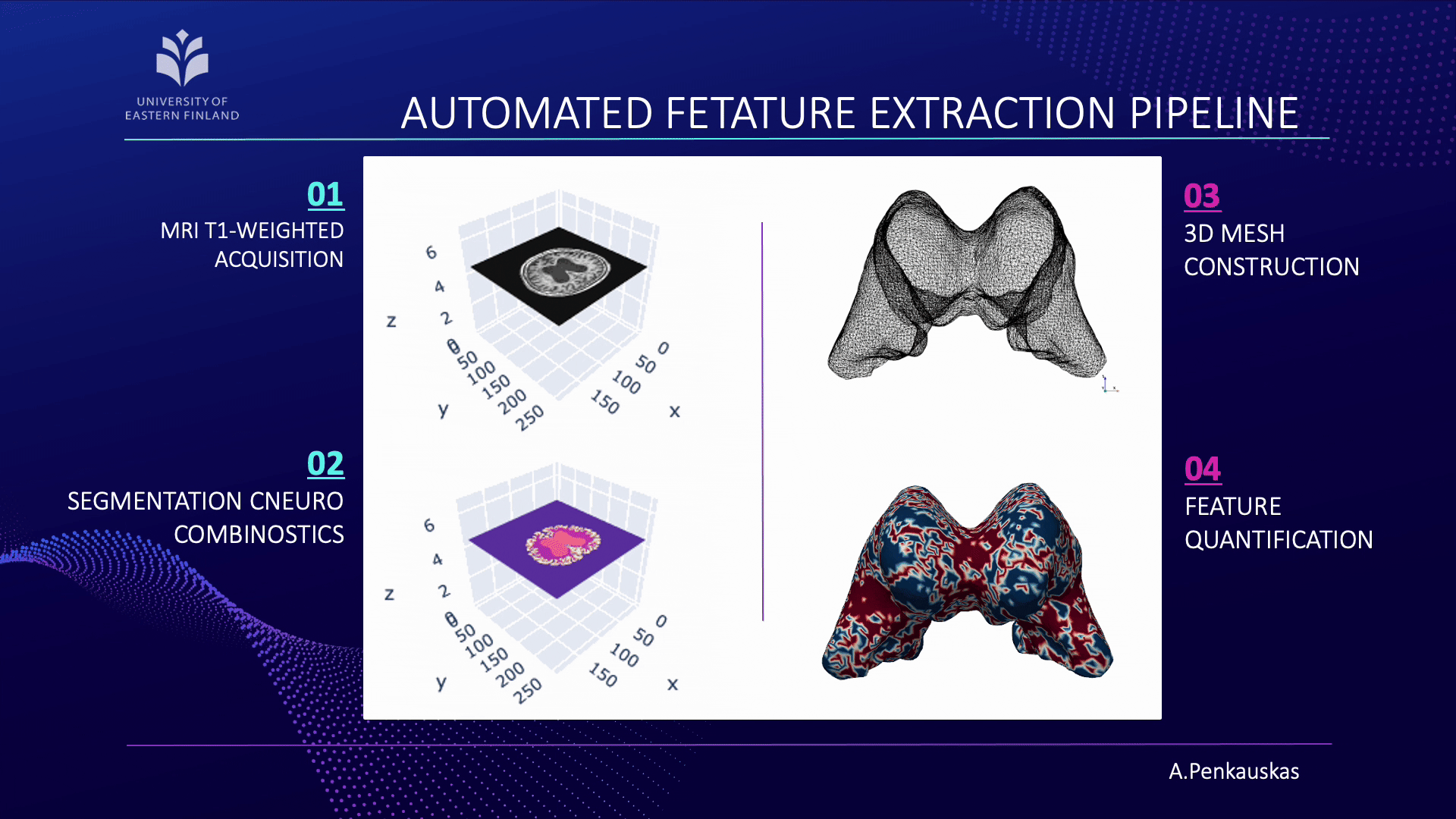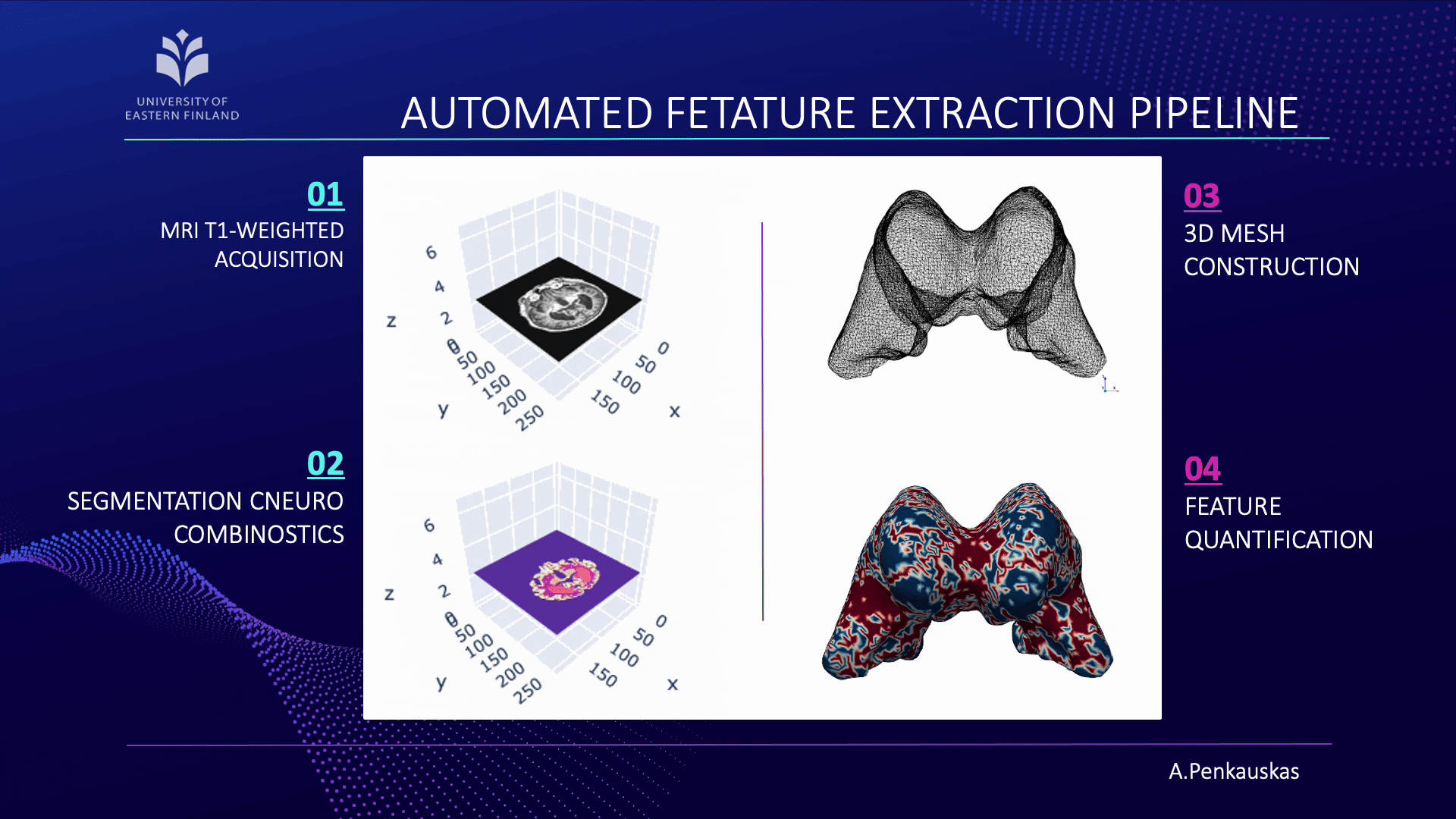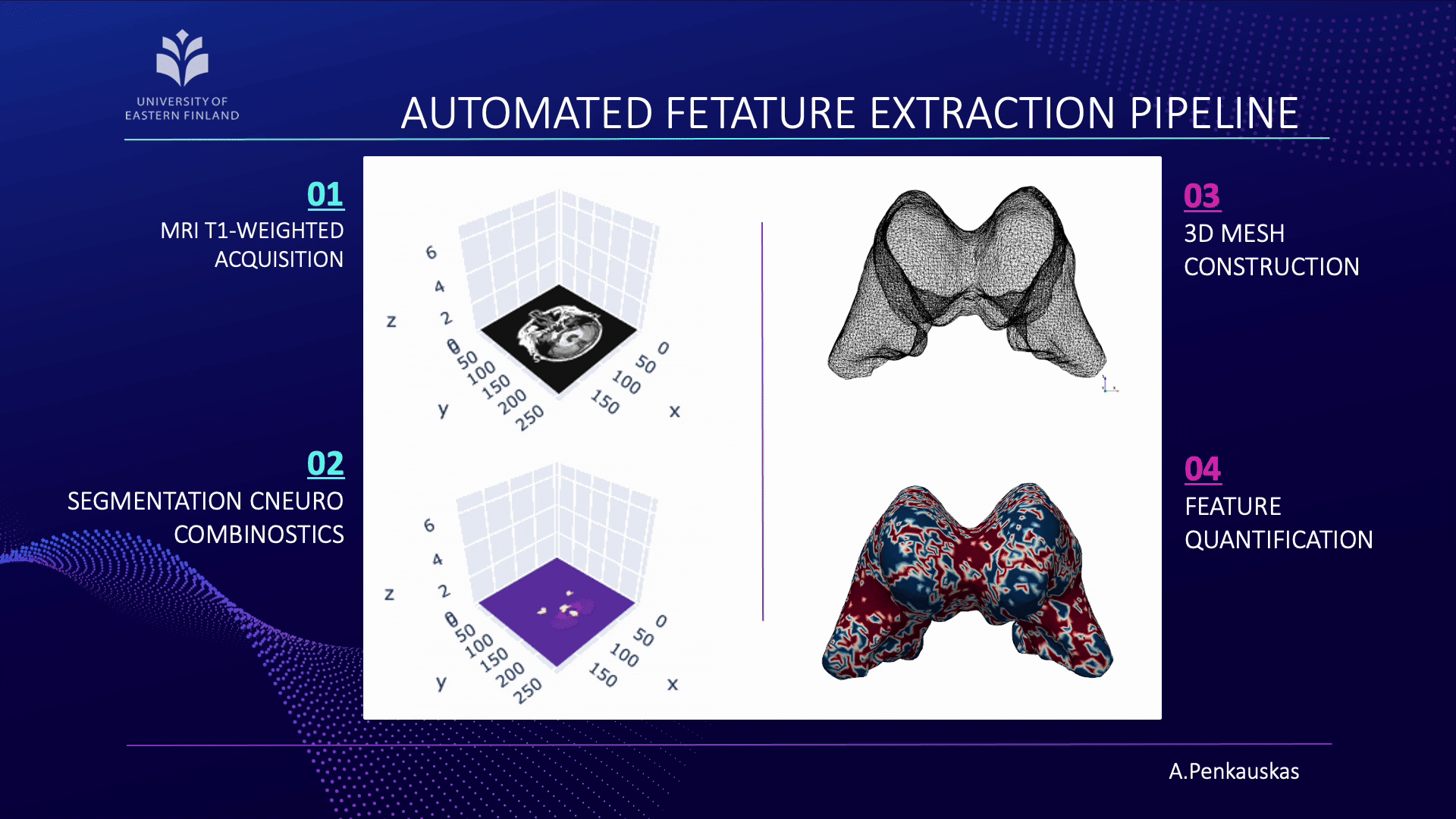
If you are curious about the process, let me briefly unravel it for you. Our workflow consists of four main steps:
- First, the acquisition of brain images using magnetic resonance imaging (MRI).
- Next, segmentation of brain scans to isolate the lateral ventricles, our primary region of interest.
- Then, the construction of three-dimensional (3D) meshes is performed from these segmented regions.
- Finally, quantification of curvature and geometric features is achieved employing 3D meshes.
When combined with demographic attributes and cerebrospinal fluid biomarkers, these measurements enable the prediction of amyloid pathology presence. This approach represents a step toward less invasive identification of patients at risk and contributes to advancing diagnostic precision.
Ultimately, it is up to you, the reader, to determine whether my explanation has meaningfully addressed the “So what?” question. Regardless of whether I have succeeded in bridging that gap today, the true purpose of my research remains clear.
My aim is to dedicate my expertise and effort to developing tools that can alleviate patient suffering, work motivated not only by scientific curiosity but also by genuine compassion for those affected.
Andrius Penkauskas works as a doctoral researcher in the Neuro-Innovation PhD Programme. His research focuses on multimodal data analysis of iNPH (a type of dementia).