People
Jaakko Pohjoismäki, Professor of Molecular Biology and Genetics, PI
While having a background in biomedicine and mitochondrial biology, it was the “endless forms most beautiful” that inspired me to study biology in the first place. I am fascinated by functional genetics in non-model organisms, species diversity, and the molecular mechanisms shaping biological variation and adaptation. I am deeply passionate about nature and its persistence in a rapidly changing world, and I see research, teaching, and public engagement as complementary tools for improving our understanding of biodiversity and informing responsible environmental decision-making.

Steffi Goffart, University Lecturer
Steffi is a molecular cell biologist who is interested in comparative functional studies of cells, with particular emphasis on mitochondrial DNA (mtDNA) maintenance, mitochondrial biogenesis, and cellular physiology. Her work aims to understand how fundamental cellular processes vary across species and contribute to adaptation and health. She is responsible for the isolation, characterization, and long-term maintenance of the established cell lines in the Cell Zoo, ensuring their quality and availability for downstream genomic and functional research.

Dr Danilo Santoro, Postdoctoral researcher
As a Postdoctoral Researcher in Evolutionary Cell Biology and Bioinformatics, I focus on those dynamics ruling the delicate balance between cell cycle regulation and metabolic processes. My main activities involve developing and applying pipelines for transcriptomics, chromatin conformation (Hi-C), and cross-species single-cell data integration. Another aspect of my research relies on assembling reference genomes and comparing closely related species to identify structural genomic differences.

MSc Ling Zhao, PhD student
I am a PhD student in the HARECY-project. I have a background in mathematics, systems science, and machine learning, and I am currently training in bioinformatics and evolutionary genomics.

MSc Jiri Vihavainen, PhD student
I’ve always been astonished how versatile and diverse group the insects are. During my bachelor studies, I started to find interest in collecting different species of Coleoptera and Orthoptera, both groups which I still keenly collect. As my taxonomic knowledge started to grow, I began to notice the flaws of relying purely on morphological cues in species identification, as every other person seemed to think a specimen was a different group than the other. Of course, we were still students at the time, but this observation stayed with me throughout my studies. With my second interest of laboratory work, especially working with DNA, I found the world of DNA barcoding, and finally I could mix my two research interests in Biodiversity Genomics.
Now in my PhD, I am working towards shedding light to our dark taxa with morphologically challenging group of flies, the scuttle flies (Diptera: Phoridae). This project focuses on developing new methods to tackle the taxonomic impediment of our vast biodiversity.

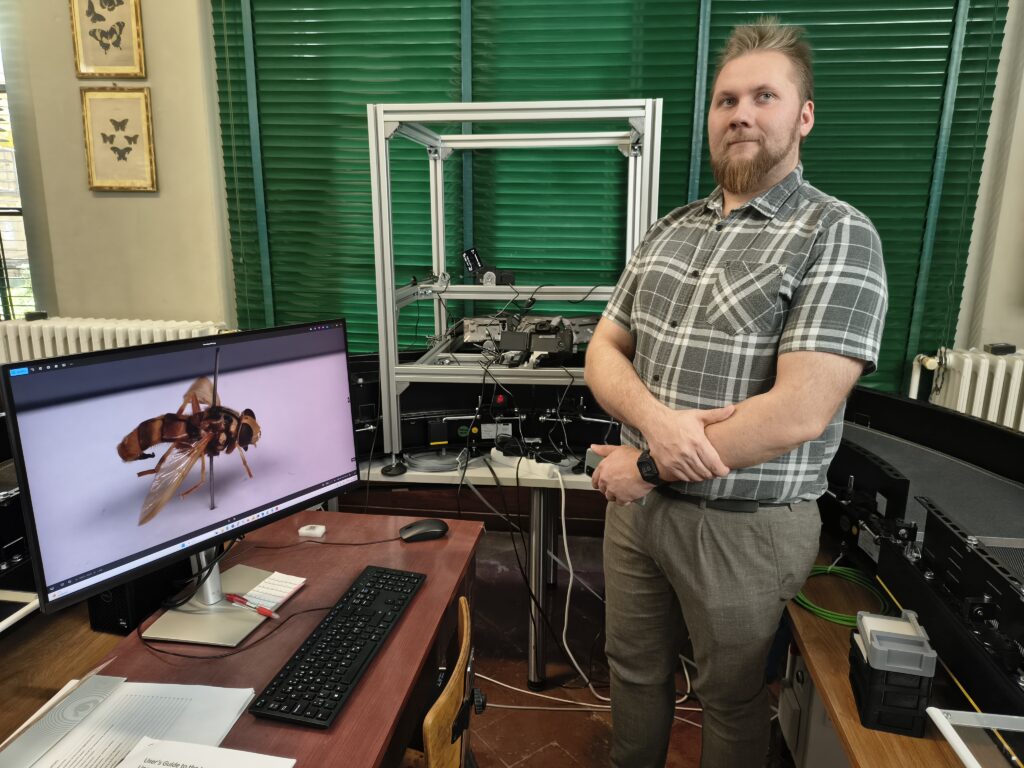
MSc Niko Lappalainen, PhD student
My research will focus on advancing automated mass digitization pipelines for natural history collections, both at the level of hardware level, with conveyor imaging and software level, with database creation. I intend to characterize the current technical limitations of conveyor-based imaging systems and optimize control configurations and quantify how design choices affect throughput and image quality in real-world institutional environments. This work will lead me to develop pipelines for automated quality control, specimen and label segmentation, multi-barcode reading, as well as deploy current OCR technologies for handwritten labels. Overall goal of this work is to achieve a better level of data extraction from natural history samples with lesser manual labour.
Although my work is not directly focused on genomics, the digitization of natural history specimens is a crucial component of modern biodiversity research and molecular ecology. In the context of DNA barcoding in particular, it is essential that each sequenced specimen is linked to a carefully documented and imaged voucher specimen to which the genetic data can be unambiguously traced. High-quality images enable later morphological examination, validation of species identifications, and reproducibility of results, even when direct access to the physical specimen is no longer possible.
Systematic and automated digitization therefore improves the reliability, interoperability, and long-term value of DNA barcode data. It supports taxonomic research, facilitates data sharing at the international level, and ensures that genetic information remains firmly connected to the physical museum specimens that form the backbone of biodiversity science.